Classification trees
Contents
7.2. Classification trees¶
Classification trees are very similar to regression trees, except that the target variable is categorical. In regression trees, we used the mean of the target variable in each region as the prediction. In classification trees, we use the most common class in each region as the prediction. Besides the most common class, we are also interested in the proportion of each class in each region. This is because the proportion of each class in each region is a measure of the purity of the region. A region is pure if it contains only one class. A region is impure if it contains multiple classes.
7.2.1. Gini index¶
Although classification accuracy is a good measure for classification performance, a popular cost function (splitting criterion) for classification trees is the Gini index, a measure of the impurity of a region (not just feature regions in machine learning, but also regions in society in economics, e.g. to measure income/wealth inequality). This is because classification accuracy is not sufficiently sensitive for building classification trees.
The Gini index is defined as:
where \(p_c\) is the proportion of class \(c\) in the region. The Gini index is zero if the region is pure, and is one if the region is impure. The Gini index is a measure of the probability of misclassification. The Gini index is also known as the Gini impurity.
7.2.2. Entropy¶
Another popular cost function (splitting criterion) for classification trees is the entropy, which is defined as:
where \(p_c\) is the proportion of class \(c\) in the region. The entropy is zero if the region is pure, and is one if the region is impure.
When building classification trees, either the Gini index or the entropy are typically used to evaluate the quality of a particular split, and the split that produces the lowest cost is chosen. The Gini index and the entropy are very similar, and the Gini index is slightly faster to compute.
7.2.3. Classification trees for heart disease diagnosis¶
Get ready by importing the APIs needed from respective libraries.
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier, plot_tree
from sklearn.metrics import confusion_matrix, accuracy_score, classification_report
import matplotlib.pyplot as plt
%matplotlib inline
Set a random seed for reproducibility.
np.random.seed(2022)
We use the Heart dataset (click to explore) to predict whether a patient has heart disease or not. The target variable is AHD, which is a binary variable that indicates whether a patient has heart disease or not.
Load the dataset and remove the rows with missing values. Inspect the first few rows of the dataset.
heart_url = "https://github.com/pykale/transparentML/raw/main/data/Heart.csv"
heart_df = pd.read_csv(heart_url).dropna()
heart_df.head()
| Unnamed: 0 | Age | Sex | ChestPain | RestBP | Chol | Fbs | RestECG | MaxHR | ExAng | Oldpeak | Slope | Ca | Thal | AHD | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 63 | 1 | typical | 145 | 233 | 1 | 2 | 150 | 0 | 2.3 | 3 | 0.0 | fixed | No |
| 1 | 2 | 67 | 1 | asymptomatic | 160 | 286 | 0 | 2 | 108 | 1 | 1.5 | 2 | 3.0 | normal | Yes |
| 2 | 3 | 67 | 1 | asymptomatic | 120 | 229 | 0 | 2 | 129 | 1 | 2.6 | 2 | 2.0 | reversable | Yes |
| 3 | 4 | 37 | 1 | nonanginal | 130 | 250 | 0 | 0 | 187 | 0 | 3.5 | 3 | 0.0 | normal | No |
| 4 | 5 | 41 | 0 | nontypical | 130 | 204 | 0 | 2 | 172 | 0 | 1.4 | 1 | 0.0 | normal | No |
Take a look at the structure of the dataset.
heart_df.info()
<class 'pandas.core.frame.DataFrame'>
Index: 297 entries, 0 to 301
Data columns (total 15 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 297 non-null int64
1 Age 297 non-null int64
2 Sex 297 non-null int64
3 ChestPain 297 non-null object
4 RestBP 297 non-null int64
5 Chol 297 non-null int64
6 Fbs 297 non-null int64
7 RestECG 297 non-null int64
8 MaxHR 297 non-null int64
9 ExAng 297 non-null int64
10 Oldpeak 297 non-null float64
11 Slope 297 non-null int64
12 Ca 297 non-null float64
13 Thal 297 non-null object
14 AHD 297 non-null object
dtypes: float64(2), int64(10), object(3)
memory usage: 37.1+ KB
Use the factorize method from the pandas library to convert categorical variables to numerical variables. This is because the sklearn library only accepts numerical variables. ChestPain is a categorical variable that indicates the type of chest pain. Thal is a categorical variable that indicates the type of thalassemia.
heart_df.ChestPain = pd.factorize(heart_df.ChestPain)[0]
heart_df.Thal = pd.factorize(heart_df.Thal)[0]
Get the feature matrix X and the target vector y.
X = heart_df.drop("AHD", axis=1)
y = pd.factorize(heart_df.AHD)[0]
Fit a classification tree to the data using the DecisionTreeClassifier class from the sklearn.tree module. Set the max_leaf_nodes parameter to 6. This will limit the number of leaf nodes to 6. Set the max_features parameter to 3. This will limit the number of features to consider when looking for the best split to 3. It will randomly select these 3 features at each split.
clf = DecisionTreeClassifier(max_leaf_nodes=6, max_features=3)
clf.fit(X, y)
DecisionTreeClassifier(max_features=3, max_leaf_nodes=6)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
DecisionTreeClassifier(max_features=3, max_leaf_nodes=6)
Compute the accuracy of the classification tree on the training data.
clf.score(X, y)
0.8047138047138047
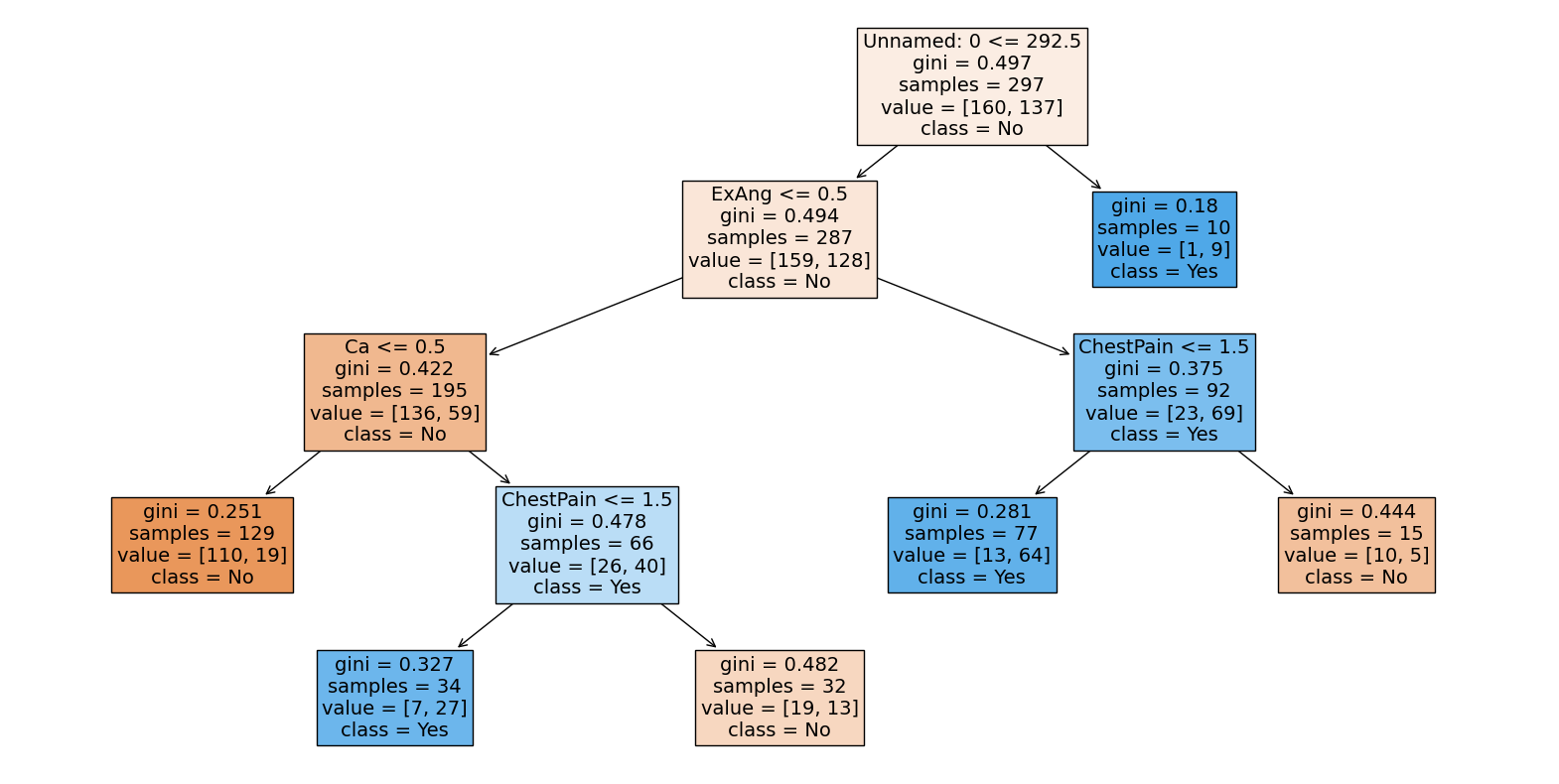
Visualise the classification tree using the plot_tree function from the sklearn.tree module. Set the filled parameter to True to colour the nodes in the tree according to the majority class in each region. Set the feature_names parameter to the list of feature names. Set the class_names parameter to the list of class names.
plt.figure(figsize=(20, 10))
plot_tree(
clf, filled=True, feature_names=X.columns, class_names=["No", "Yes"], fontsize=14
)
plt.show()
7.2.4. Classification trees for car seat sales¶
Next, let us study the Carseats dataset (click to explore). In this dataset, we want to predict whether a car seat will be High or Low based on the Sales and Price of the car seat.
Load the dataset. Sales is a continuous variable so we recode it as a binary variable High by thresholding it at 8 using the map() function from the pandas library. High takes on a value of ‘Y’ if the Sales variable exceeds 8, ‘N’ otherwise. We also convert categorical variables to numerical variables using the factorize method from the pandas library as above.
Inspect the first few rows of the dataset.
carseats_url = "https://github.com/pykale/transparentML/raw/main/data/Carseats.csv"
carseats_df = pd.read_csv(carseats_url)
carseats_df["High"] = carseats_df.Sales.map(lambda x: "Y" if x > 8 else "N")
carseats_df.ShelveLoc = pd.factorize(carseats_df.ShelveLoc)[0]
carseats_df.Urban = carseats_df.Urban.map(lambda x: 1 if x == "Yes" else 0)
carseats_df.US = carseats_df.US.map(lambda x: 1 if x == "Yes" else 0)
carseats_df.head()
| Sales | CompPrice | Income | Advertising | Population | Price | ShelveLoc | Age | Education | Urban | US | High | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 9.50 | 138 | 73 | 11 | 276 | 120 | 0 | 42 | 17 | 1 | 1 | Y |
| 1 | 11.22 | 111 | 48 | 16 | 260 | 83 | 1 | 65 | 10 | 1 | 1 | Y |
| 2 | 10.06 | 113 | 35 | 10 | 269 | 80 | 2 | 59 | 12 | 1 | 1 | Y |
| 3 | 7.40 | 117 | 100 | 4 | 466 | 97 | 2 | 55 | 14 | 1 | 1 | N |
| 4 | 4.15 | 141 | 64 | 3 | 340 | 128 | 0 | 38 | 13 | 1 | 0 | N |
Take a look at the structure of the dataset.
carseats_df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 400 entries, 0 to 399
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Sales 400 non-null float64
1 CompPrice 400 non-null int64
2 Income 400 non-null int64
3 Advertising 400 non-null int64
4 Population 400 non-null int64
5 Price 400 non-null int64
6 ShelveLoc 400 non-null int64
7 Age 400 non-null int64
8 Education 400 non-null int64
9 Urban 400 non-null int64
10 US 400 non-null int64
11 High 400 non-null object
dtypes: float64(1), int64(10), object(1)
memory usage: 37.6+ KB
Split the dataset into training (200 samples) and test sets.
X2 = carseats_df.drop(["Sales", "High"], axis=1)
y2 = carseats_df.High
train_size = 200
X_train, X_test, y_train, y_test = train_test_split(
X2, y2, train_size=train_size, test_size=X.shape[0] - train_size
)
For this dataset, let us use ‘entropy’ as the split criterion at each node to fit a classification tree to the training data.
criteria = "entropy"
max_depth = 6
min_sample_leaf = 4
clf_gini = DecisionTreeClassifier(
criterion=criteria, max_depth=max_depth, min_samples_leaf=min_sample_leaf
)
clf_gini.fit(X_train, y_train)
print(clf_gini.score(X_train, y_train))
0.895
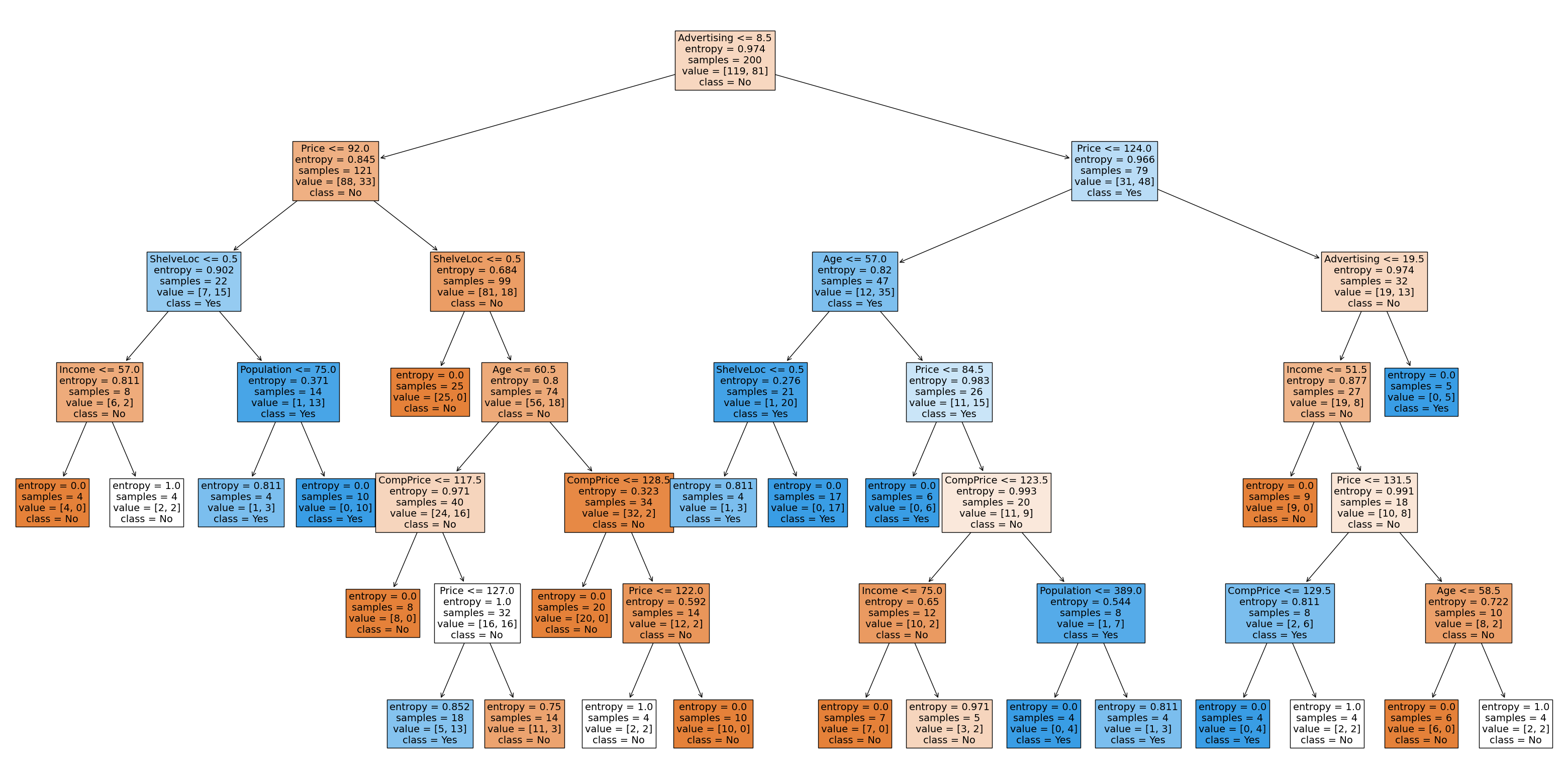
Visualise the classification tree.
# one attractive feature of a tree is visulization.
plt.figure(figsize=(40, 20)) # customize according to the size of your tree
plot_tree(
clf_gini,
filled=True,
feature_names=X_train.columns,
class_names=["No", "Yes"],
fontsize=14,
)
plt.show()
Build the confusion matrix to evaluate the model in accuracy for both training and test datasets.
y_pred_train = clf_gini.predict(X_train)
cm = pd.DataFrame(
confusion_matrix(y_train, y_pred_train).T,
index=["No", "Yes"],
columns=["No", "Yes"],
)
print(cm)
print("Train Accuracy is ", accuracy_score(y_train, y_pred_train) * 100)
y_pred = clf_gini.predict(X_test)
cm = pd.DataFrame(
confusion_matrix(y_test, y_pred).T, index=["No", "Yes"], columns=["No", "Yes"]
)
print(cm)
print("Test Accuracy is ", accuracy_score(y_test, y_pred) * 100)
No Yes
No 111 13
Yes 8 68
Train Accuracy is 89.5
No Yes
No 46 15
Yes 13 23
Test Accuracy is 71.1340206185567
The test accuracy of the fitted model is significantly lower than the training accuracy, an indication of overfitting. You can go back and change the hyperparameters in the training process to reduce the dimension (flexibility) of the parameter space and reduce overfitting.
Compute more metrics such as precision, recall, f1-score, etc.
print(classification_report(y_test, y_pred))
precision recall f1-score support
N 0.75 0.78 0.77 59
Y 0.64 0.61 0.62 38
accuracy 0.71 97
macro avg 0.70 0.69 0.69 97
weighted avg 0.71 0.71 0.71 97
7.2.5. Exercises¶
1. All the following exercises use the Caravan dataset.
Load the Caravan dataset and inspect the first five rows.
# Write your code below to answer the question
Compare your answer with the reference solution below
import pandas as pd
caravan_url = "https://github.com/pykale/transparentML/raw/main/data/Caravan.csv"
caravan_df = pd.read_csv(caravan_url)
caravan_df.head()
| MOSTYPE | MAANTHUI | MGEMOMV | MGEMLEEF | MOSHOOFD | MGODRK | MGODPR | MGODOV | MGODGE | MRELGE | ... | APERSONG | AGEZONG | AWAOREG | ABRAND | AZEILPL | APLEZIER | AFIETS | AINBOED | ABYSTAND | Purchase | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 33 | 1 | 3 | 2 | 8 | 0 | 5 | 1 | 3 | 7 | ... | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | No |
| 1 | 37 | 1 | 2 | 2 | 8 | 1 | 4 | 1 | 4 | 6 | ... | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | No |
| 2 | 37 | 1 | 2 | 2 | 8 | 0 | 4 | 2 | 4 | 3 | ... | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | No |
| 3 | 9 | 1 | 3 | 3 | 3 | 2 | 3 | 2 | 4 | 5 | ... | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | No |
| 4 | 40 | 1 | 4 | 2 | 10 | 1 | 4 | 1 | 4 | 7 | ... | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | No |
5 rows × 86 columns
2. Now, split the Caravan dataset into train and test sets using the proportion 80:20 and fit a DecisionTreeClassifier with \(8\) maximum leaf nodes and \(6\) maximum features. Calculate the testing accuracy and AUC of the fitted model. (Use \(2022\) as the random seed value)
# Write your code below to answer the question
Compare your answer with the reference solution below
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier, plot_tree
from sklearn.metrics import accuracy_score, roc_auc_score
np.random.seed(2022)
X2 = caravan_df.drop(["Purchase"], axis=1)
y2 = caravan_df.Purchase
X_train, X_test, y_train, y_test = train_test_split(X2, y2, test_size=0.20)
clf = DecisionTreeClassifier(max_leaf_nodes=8, max_features=6, random_state=2022)
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
accuarcy = accuracy_score(y_test, y_pred)
print("Accuracy: ", accuarcy)
y_prob = clf.predict_proba(X_test)[:, 1]
roc_auc = roc_auc_score(y_test, y_prob)
print("AUC: ", roc_auc)
Accuracy: 0.9399141630901288
AUC: 0.5891454664057404
3. Now, train another DecisionTreeClassifier using entropy as the split criterion at each node to fit a classification tree with a maximum depth of \(4\) and a minimum sample leaf of \(3\) on the training data and find out the testing accuracy and AUC. (Use \(2022\) as the random seed value)
# Write your code below to answer the question
Compare your answer with the reference solution below
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier, plot_tree
np.random.seed(2022)
criteria = "entropy"
max_depth = 4
min_sample_leaf = 3
clf_gini = DecisionTreeClassifier(
criterion=criteria, max_depth=max_depth, min_samples_leaf=min_sample_leaf
)
clf_gini.fit(X_train, y_train)
y_pred = clf_gini.predict(X_test)
accuarcy = accuracy_score(y_test, y_pred)
print("Accuracy: ", accuarcy)
y_prob = clf_gini.predict_proba(X_test)[:, 1]
roc_auc = roc_auc_score(y_test, y_prob)
print("AUC: ", roc_auc)
Accuracy: 0.9399141630901288
AUC: 0.7402804957599478
4. Compare the performance of the trained models in Exercise 3 with Exercise 2.
Compare your answer with the solution below
In terms of testing accuracy, the Exercise 2 model outperformed the Exercise 3 model, but accuracy is not the only metric to evaluate the models. When we look at the AUC score, which more comprehensively evaluates a model in imbalanced tasks, the Exercise 3 model outperforms the Exercise 2 model. Therefore, in this case we can see the Exercise 3 model is better than the Exercise 2 model.
5. Create a confusion matrix and a classification report to help you evaluate the model you trained in Exercise 3.
# Write your code below to answer the question
Compare your answer with the reference solution below
from sklearn.metrics import confusion_matrix, classification_report
y_pred = clf_gini.predict(X_test)
cm = pd.DataFrame(
confusion_matrix(y_test, y_pred).T, index=["No", "Yes"], columns=["No", "Yes"]
)
print("Confusion matrix\n", cm)
print(classification_report(y_test, y_pred))
Confusion matrix
No Yes
No 1093 68
Yes 2 2
precision recall f1-score support
No 0.94 1.00 0.97 1095
Yes 0.50 0.03 0.05 70
accuracy 0.94 1165
macro avg 0.72 0.51 0.51 1165
weighted avg 0.91 0.94 0.91 1165
a. What is the testing accuracy of the model trained in Exercise 3?
Compare your answer with the solution below
The testing accuracy of the model trained in Exercise 3 is 0.94.
b. Does the model predicted well?
Compare your answer with the solution below
If we only consider the test accuracy, we may conclude that the model learned the task effectively, but this is not the whole story. In fact, due to the class imbalance in the training data, this model is biased towards the “NO” class. If we look at the confusion matrix, we see that it predicts “NO” for almost all samples, and has a poor recall and precision rate for the “YES” class. Again, this shows that accuracy alone is also not always a good metric for evaluating models. Considering AUC, recall, and precision as well as showing the confusion matrix, we can get a much better picture.
